MSL-IO
MSL-IO follows the data model used by HDF5 to read and write data files – where there is a
Root, Groups and Datasets and these objects
each have Metadata associated with them.
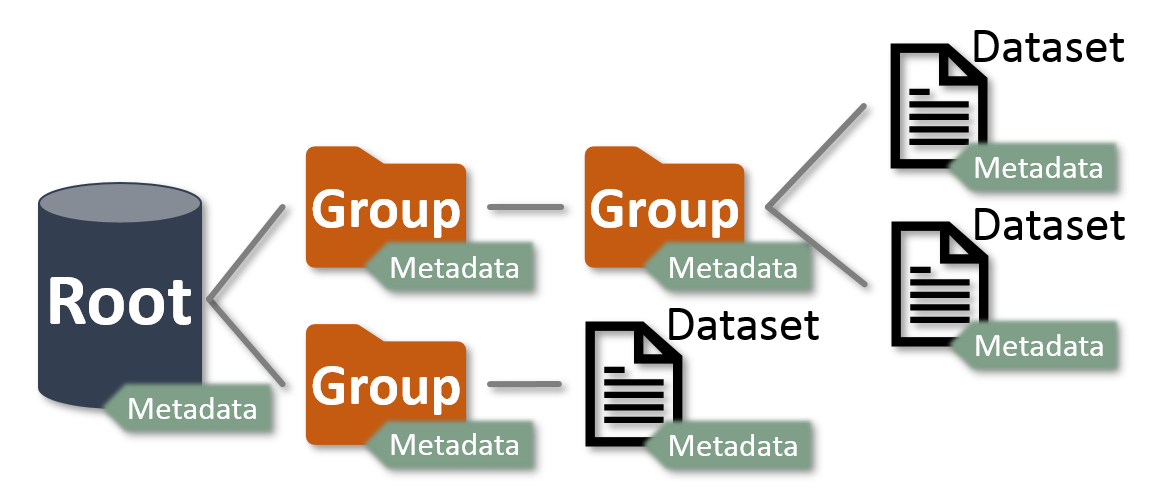
The tree structure is similar to the file-system structure used by operating systems. Groups
are analogous to the directories (where Root is the root Group) and
Datasets are analogous to the files.
The data files that can be read or created are not restricted to HDF5 files, but any file format that has a Reader implemented can be read and data files can be created using any of the Writers.
Getting Started
Write a file
Suppose you want to create a new HDF5 file. We first create an instance of
HDF5Writer
>>> from msl.io import HDF5Writer
>>> h5 = HDF5Writer()
then we can add Metadata to the Root,
>>> h5.add_metadata(one=1, two=2)
>>> dataset1 = h5.create_dataset('dataset1', data=[1, 2, 3, 4])
>>> my_group = h5.create_group('my_group')
and create a Dataset in my_group
>>> dataset2 = my_group.create_dataset('dataset2', data=[[1, 2], [3, 4]], three=3)
Finally, we write the file
>>> h5.write(file='my_file.h5')
Read a file
The read() function is available to read a file. Provided that a Reader
exists to read the file a Root object is returned. We will read the file
that we created above.
>>> from msl.io import read
>>> root = read('my_file.h5')
You can print a representation of all Groups and Datasets
in the Root by calling the tree() method
>>> print(root.tree())
<HDF5Reader 'my_file.h5' (1 groups, 2 datasets, 2 metadata)>
<Dataset '/dataset1' shape=(4,) dtype='<f8' (0 metadata)>
<Group '/my_group' (0 groups, 1 datasets, 0 metadata)>
<Dataset '/my_group/dataset2' shape=(2, 2) dtype='<f8' (1 metadata)>
Since the root object is a Group (which operates like a Python dict) you can
iterate over the items that are in the file using
>>> for name, value in root.items():
... print('{!r} -- {!r}'.format(name, value))
'/dataset1' -- <Dataset '/dataset1' shape=(4,) dtype='<f8' (0 metadata)>
'/my_group' -- <Group '/my_group' (0 groups, 1 datasets, 0 metadata)>
'/my_group/dataset2' -- <Dataset '/my_group/dataset2' shape=(2, 2) dtype='<f8' (1 metadata)>
where value will either be a Group or a Dataset.
You can iterate over the Groups that are in the file
>>> for group in root.groups():
... print(group)
<Group '/my_group' (0 groups, 1 datasets, 0 metadata)>
or iterate over the Datasets
>>> for dataset in root.datasets():
... print(repr(dataset))
<Dataset '/dataset1' shape=(4,) dtype='<f8' (0 metadata)>
<Dataset '/my_group/dataset2' shape=(2, 2) dtype='<f8' (1 metadata)>
You can access the Metadata of any object through the metadata attribute
>>> root.metadata
<Metadata '/' {'one': 1, 'two': 2}>
You can access values of the Metadata as attributes
>>> root.metadata.one
1
>>> dataset2.metadata.three
3
or as keys
>>> root.metadata['two']
2
>>> dataset2.metadata['three']
3
When root is returned it is accessed in read-only mode
>>> root.read_only
True
>>> for name, value in root.items():
... print('is {!r} in read-only mode? {}'.format(name, value.read_only))
is '/dataset1' in read-only mode? True
is '/my_group' in read-only mode? True
is '/my_group/dataset2' in read-only mode? True
If you want to edit the Metadata for root, or modify any
Groups or Datasets in root, then you must first set
the object to be editable. Setting the read-only mode of root propagates that mode to all items within
root. For example,
>>> root.read_only = False
will make root and all Groups and all Datasets within root to be editable
>>> root.read_only
False
>>> for name, value in root.items():
... print('is {!r} in read-only mode? {}'.format(name, value.read_only))
is '/dataset1' in read-only mode? False
is '/my_group' in read-only mode? False
is '/my_group/dataset2' in read-only mode? False
You can make only a specific object (and it’s descendants) editable as well. You can make
my_group and dataset2 to be in read-only mode by the following (recall that root behaves
like a Python dict)
>>> root['my_group'].read_only = True
and this will keep root and dataset1 in editable mode, but change my_group and dataset2 to be in read-only mode
>>> root.read_only
False
>>> for name, value in root.items():
... print('is {!r} in read-only mode? {}'.format(name, value.read_only))
is '/dataset1' in read-only mode? False
is '/my_group' in read-only mode? True
is '/my_group/dataset2' in read-only mode? True
You can access the Groups and Datasets as keys or as class attributes
>>> root['my_group']['dataset2'].shape
(2, 2)
>>> root.my_group.dataset2.shape
(2, 2)
See Accessing Keys as Class Attributes for more information.
Convert a file
You can convert between file formats using any of the Writers. Suppose you had an HDF5 file and you wanted to convert it to the JSON format
>>> from msl.io import JSONWriter
>>> h5 = read('my_file.h5')
>>> writer = JSONWriter('my_file.json')
>>> writer.write(root=h5)
Read data in a table
The read_table() function is available to read a table from a file.
A table has the following properties:
The first row is a header.
All rows have the same number of columns.
All data values in a column have the same data type.
The returned object is a Dataset with the header provided as metadata.
Suppose a file called my_table.csv contains the following information
| x, | y, | z |
|---|---|---|
| 1, | 2, | 3 |
| 4, | 5, | 6 |
| 7, | 8, | 9 |
You can read this file and interact with the data using the following
>>> from msl.io import read_table
>>> csv = read_table('my_table.csv')
>>> csv
<Dataset 'my_table.csv' shape=(3, 3) dtype='<f8' (1 metadata)>
>>> csv.metadata
<Metadata 'my_table.csv' {'header': array(['x', 'y', 'z'], dtype='<U1')}>
>>> csv.data
array([[1., 2., 3.],
[4., 5., 6.],
[7., 8., 9.]])
>>> csv.max()
9.0
You can read a table from a text-based file or from an Excel spreadsheet.